[ad_1]
For the reason that Nineteen Fifties, scientists have had a fairly good concept of how muscle groups work. The protein on the centre of the motion is myosin, a molecular motor that ratchets itself alongside rope-like strands of actin proteins — greedy, pulling, releasing and greedy once more — to make muscle cells contract.
The fundamentals had been first defined in a pair of landmark papers in Nature1,2, they usually have been confirmed and elaborated on by detailed molecular maps of myosin and its companions. Researchers suppose that myosin generates power by cocking again the lengthy lever-like arm that’s connected to the motor portion of the protein.
The one hitch is that scientists had by no means seen this fleeting pre-stroke state — till now.
In a preprint revealed in January3, researchers used a cutting-edge structural biology method to report this second, which lasts simply milliseconds in dwelling cells.
‘Your entire protein universe’: AI predicts form of almost each recognized protein
“It’s one of many issues within the textbook you type of gloss over,” says Stephen Muench, a structural biologist on the College of Leeds, UK, who co-led the research. “These are experiments that individuals needed to do 40 years in the past, however they only by no means had the expertise.”
That expertise — referred to as time-resolved cryo-electron microscopy (cryo-EM) — now has structural biologists considering like cinematographers, turning nonetheless snapshots of life’s molecular equipment into movement photos that reveal the way it works.
Muench and his colleagues’ myosin film isn’t feature-length; it consists of simply two frames exhibiting totally different levels of the molecular movement. But it confirmed a decades-old idea and settled debates over the order of the steps in myosin’s choreography. Different researchers are focusing their new-found director’s eye on understanding cell-signalling techniques, together with these underlying opioid overdoses, the gene-editing juggernaut CRISPR–Cas9 and different molecular machines which were principally studied with extremely detailed, but static structural maps.
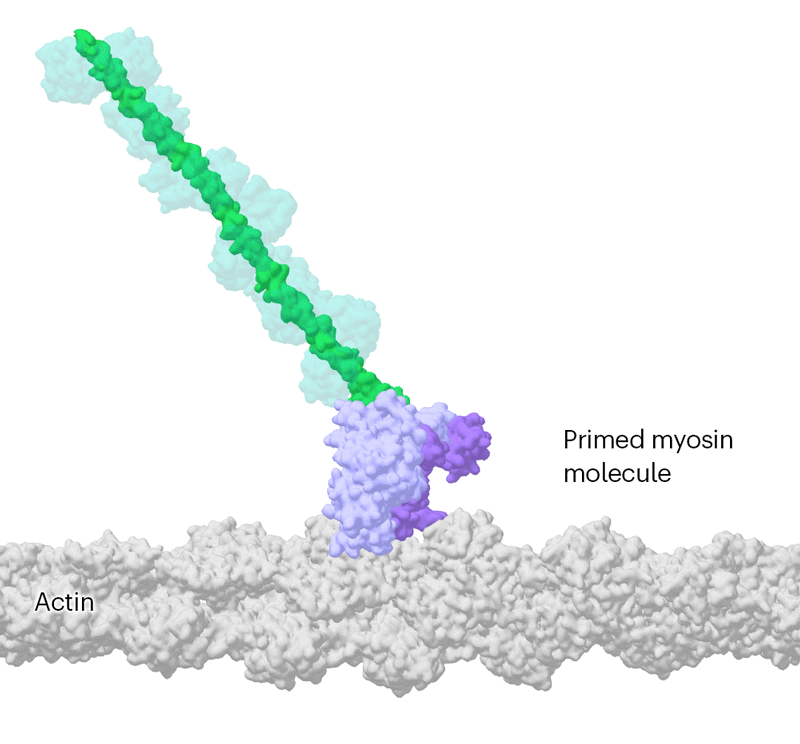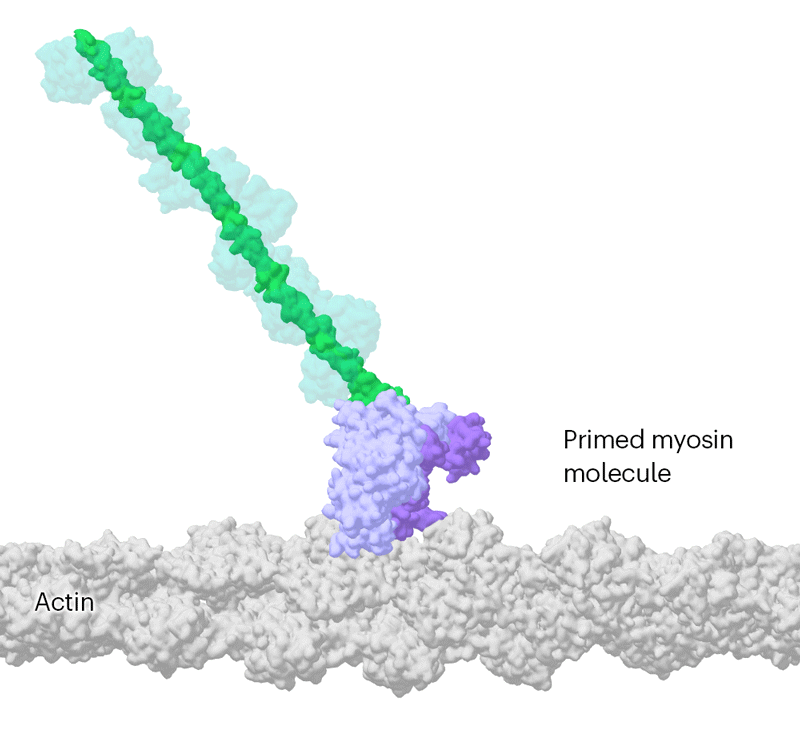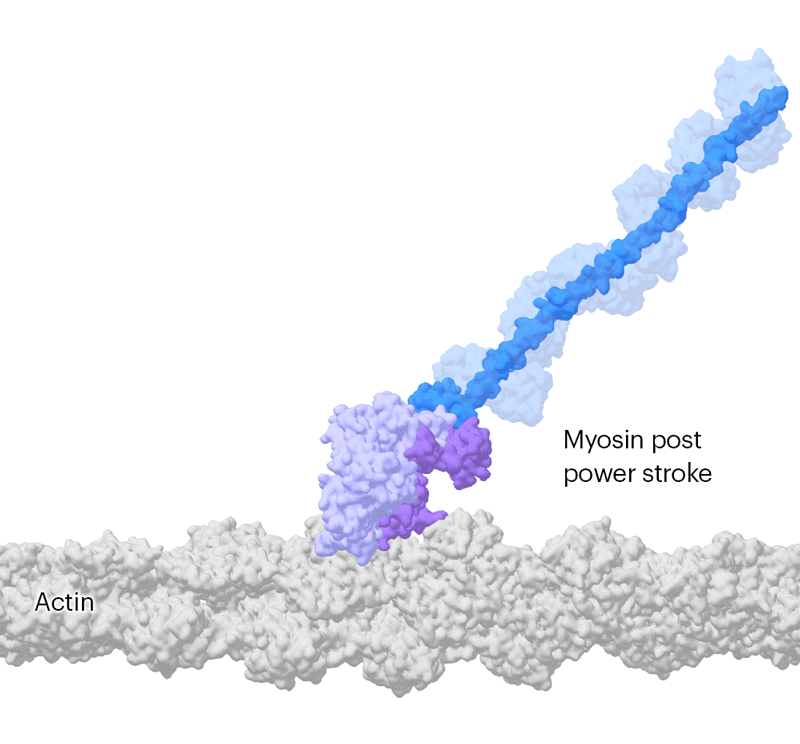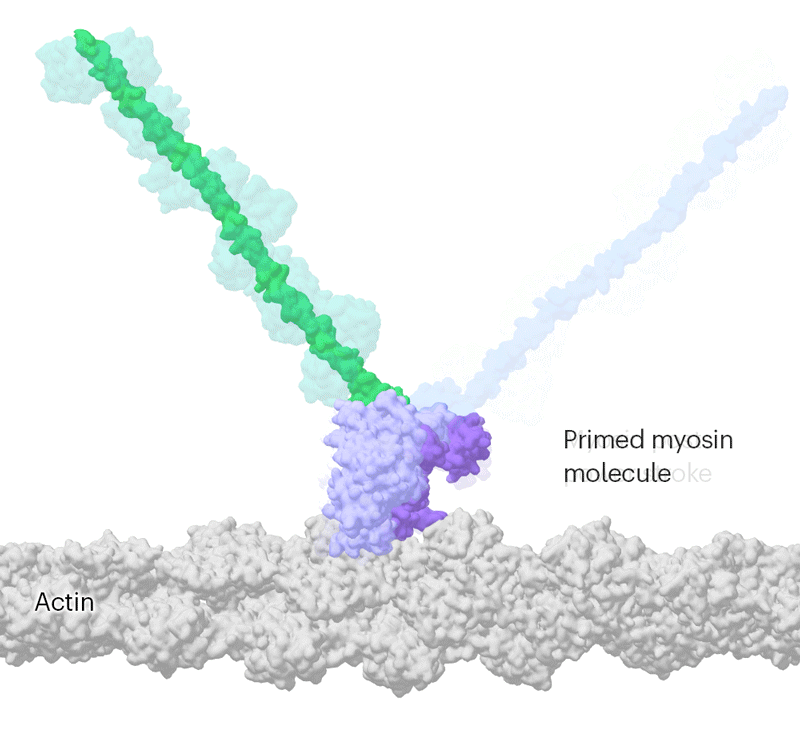
Researchers have been capable of seize photographs of particular person myosin proteins as they pull on an actin filament throughout muscle contraction, confirming key particulars of the movement. First, myosin turns into cocked or primed, then it attaches to actin and its lever arm swings in an influence stroke that slides the filament by about 34 nanometres.Credit score: Sean McMillan
“The massive image is to maneuver away, as a lot as doable, from this single, static snapshot,” says Georgios Skiniotis, a structural biologist at Stanford College in California, whose group used the method to report the activation of a kind of cell-signalling molecule referred to as a G-protein-coupled receptor (GPCR)4. “I need the film.”
Freeze body
To underscore the ability of cryo-EM, Skiniotis and others like to attract a comparability with one of many first movement photos ever made. Within the 1870s, photographer Eadweard Muybridge used high-speed images expertise, which was leading edge on the time, to seize a collection of nonetheless photographs of a galloping horse. They confirmed, for the primary time, that each one 4 of the animal’s hooves depart the bottom without delay — one thing that the human eye couldn’t distinguish.
Comparable insights, Skiniotis says, will come from making use of the identical concept to protein buildings. “I wish to get a dynamic image.”
The flexibility to map proteins and different biomolecules right down to the placement of particular person atoms has remodeled biology, underpinning advances in gene modifying, drug discovery and revolutionary artificial-intelligence instruments comparable to AlphaFold, which may predict protein buildings. However the principally static photographs delivered by X-ray crystallography and cryo-EM, the 2 applied sciences liable for the lion’s share of decided protein buildings, belie the dynamic nature of life’s molecules.
“Biomolecules are usually not made up of rocks,” says Sonya Hanson, a computational biophysicist on the Flatiron Institute in New York Metropolis. They exist in water and are consistently in movement. “They’re extra like jelly,” provides Muench.
The key lives of cells — as by no means seen earlier than
Biologists typically say that “construction determines perform”, however that’s not fairly proper, says Ulrich Lorenz, a molecular physicist on the Swiss Federal Institute of Expertise in Lausanne (EPFL). The protein poses captured by most structural research are energetically secure ‘equilibrium’ states that present restricted clues to the short-lived, unstable confirmations which can be key to chemical reactions and different features carried out by molecular machines. “Construction lets you infer perform, however solely incompletely and imperfectly, and also you’re lacking the entire particulars,” says Lorenz.
Cryo-EM is a good way to get on the particulars, however capturing these fleeting states requires cautious preparation. Protein samples are pipetted onto a grid after which flash frozen with liquid ethane. They’re then imaged utilizing highly effective electron beams that report snapshots of particular person molecules (subtle software program classifies and morphs these photos into structural maps). The samples swim in water earlier than being frozen, so any chemical response that may occur in a take a look at tube can, in idea, be frozen in place on a cryo-EM grid — if researchers can catch it rapidly sufficient.
That’s one of many first huge challenges says Joachim Frank, a structural biologist at Columbia College in New York Metropolis who shared the 2017 Nobel Prize in Chemistry for his work on cryo-EM. “Even for very dexterous folks, it takes a number of seconds.” In that point, any chemical reactions — and the intermediate buildings that mediate the reactions — could be lengthy gone earlier than freezing. “That is the hole we wish to fill,” says Frank.
Caught in translation
Frank’s group has tried to resolve this downside utilizing a microfluidic chip. The machine rapidly mixes two protein options, permits them to react for a specified time interval after which delivers response droplets onto a cryo-EM grid that’s immediately frozen.
This yr, Frank’s group used their machine to review a bacterial enzyme that rescues ribosomes, the cell’s protein-making factories, in the event that they stall in response to antibiotics or different stresses. The enzyme, referred to as HflX, helps to recycle caught ribosomes by popping their two subunits aside.
Frank’s group captured three photographs of HflX certain to the ribosome, over a span of 140 milliseconds, which present the way it splits the ribosome like somebody rigorously eradicating the shell from an oyster. The enzyme breaks a dozen or so molecular bridges that maintain a ribosome’s two subunits collectively, one after the other, till simply two are left and the ribosome pops open5. “Probably the most shocking factor to me is that it’s a really orderly course of,” Frank says. “You’d suppose the ribosome is being break up and that’s it.”
Muench and his colleagues, together with Charlie Scarff, a structural biologist on the College of Leeds, and Howard White, a kineticist at Jap Virginia Medical College in Norfolk, Virginia additionally used a microfluidic chip to make their myosin film by rapidly mixing myosin and actin3.
‘It should change all the pieces’: DeepMind’s AI makes gigantic leap in fixing protein buildings
However the molecular motor is so quick that, to gradual issues down ever additional, they used a mutated model of myosin that operates about ten instances slower than regular. This allowed the group to find out two buildings, 110 milliseconds aside, that confirmed the swing of myosin’s lever-like arm. The buildings additionally confirmed {that a} by-product of the chemical response that powers the motor — the breakdown of a mobile gasoline referred to as ATP — exits the protein’s lively website earlier than the lever swings and never after. “That’s ending many years of conjecture,” says Scarff.
With this new mannequin in thoughts, Scarff, whose specialty is myosin, and Muench are planning to make use of time-resolved cryo-EM to review how myosin dynamics are affected by sure medicine and mutations which can be recognized to trigger coronary heart illness.
Microfluidic chips aren’t the one manner researchers are placing time stamps on protein buildings. A group led by Bridget Carragher, a structural biologist and the technical director on the Chan Zuckerberg Imaging Institute in Redwood Metropolis, California, developed a ‘spray and blend’ method that entails capturing tiny volumes of reacting samples onto a grid earlier than flash-freezing them6.
In one other set-up — developed by structural physiologist Edward Twomey at Johns Hopkins College College of Drugs in Baltimore, Maryland, and his group — a flash of sunshine triggers light-sensitive chemical reactions, that are stopped by flash-freezing7. Lorenz’s equipment, in the meantime, takes already frozen samples and makes use of laser pulses to reanimate them for a number of microseconds earlier than they refreeze, all underneath the gaze of an electron microscope8.
‘Limitations all over the place’
The totally different approaches have their professionals and cons. Carragher’s spray and blend method makes use of minute pattern volumes, which ought to be straightforward to acquire for many proteins; Twomey says his ‘open-source’ light-triggered machine is comparatively cheap and may be constructed for a number of thousand {dollars}; and Lorenz says his laser-pulse system has the potential to report many extra fleeting occasions than different time-resolved cryo-EM applied sciences — right down to a tenth of a microsecond.
Revolutionary cryo-EM is taking up structural biology
However these strategies are usually not but able to be rolled out. Presently, there are not any industrial suppliers of time-resolved cryo-EM expertise, limiting its attain, says Rouslan Efremov, a structural biologist on the VIB-VUB Middle for Structural Biology in Brussels. “All these items are fussy and arduous to manage they usually haven’t actually caught on,” provides Carragher.
Holger Stark, a structural biologist on the Max Planck Institute for Multidisciplinary Sciences in Göttingen, Germany, says that present types of time-resolved cryo-EM could be helpful for some molecular machines that function on the premise of large-scale actions — for instance, the ribosome. Nevertheless, the expertise isn’t prepared to be used on simply any organic system. “You must cherry choose your topic,” he says. “We have now limitations all over the place.”
Regardless of the shortcomings, there are many fascinating questions for researchers to start out addressing now utilizing these strategies. Twomey is utilizing time-resolved cryo-EM to review Cas9, the DNA-cutting enzyme behind CRISPR gene modifying, and says the insights may assist to make extra environment friendly gene-editing techniques.
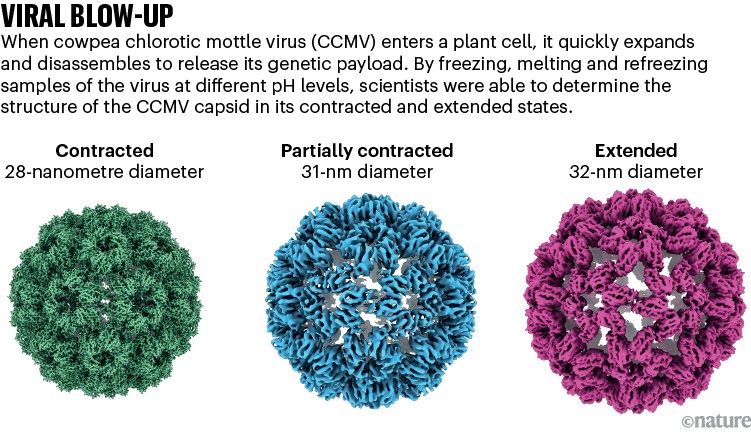
Lorenz used his laser-melting methodology to point out how a plant virus swells up after it infects a cell to launch its genetic materials7 (see ‘Viral blow-up’). He’s now learning different viral entry molecules comparable to HIV’s envelope protein. “We have now these static buildings, however we don’t know the way the system makes it from one state to the opposite, and the way the equipment works,” he says.
Supply: Ref.8
Skiniotis’s group is investigating GPCRs, together with one referred to as the β-adrenergic receptor, which has been implicated in bronchial asthma. Their work4 reveals how activating the receptor triggers it to shed its companion G-protein, a key step in propagating alerts in cells.
The researchers are actually learning the identical course of in a GPCR referred to as the µ-opioid receptor, which is activated by morphine and fentanyl amongst different medicine. In preliminary unpublished outcomes, they’ve discovered that the dynamics of the receptor assist to clarify why some medicine comparable to fentanyl are so potent in selling G-protein activation, whereas others aren’t. Such insights, says Skiniotis, are glimpses of unseen biology that molecular films promise to disclose. Simply don’t overlook the popcorn.
[ad_2]
Supply hyperlink




